Competition on International Aneurysm Segmentation
Welcome to the ICPR 2024-Competition on Intracranial Aneurysm Segmentation. This is a medical image analysis challenge organised as part of ICPR 2024.

This challenge aims to compare methods of automatically segmenting intracranial aneurysms from Time of Flight Magnetic Resonance Angiographs (TOF-MRAs).
Teams that wish to participate in this challenge should register Register here. Once your registration has been approved by the organiser, it is possible to download all of the provided training data. The training data comprises 40 sets of brain MRA images where each set includes one Time of Flight MRA (TOF- MRA) and manual annotations. All manual annotations were made by experts in the field of medical annotations and checked by an experienced radiologist.
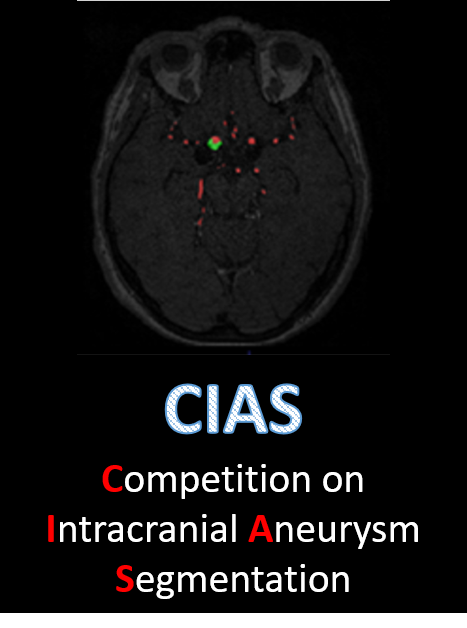
Intracranial aneurysms (IA) occur in approximately 3% of the general population, with certain demographics exhibiting elevated susceptibility. The rupture of these aneurysms leads to subarachnoid hemorrhage, causing bleeding within the brain. Timely identification of intracranial aneurysms, coupled with precise measurement and shape evaluation, holds paramount importance in clinical practice. Such measures facilitate vigilant monitoring of aneurysm growth and rupture potential, thereby facilitating well-informed treatment decisions. The objective of this endeavor is to automate the segmentation of intracranial aneurysms from Time-of-Flight Magnetic Resonance Angiography (TOF-MRA) images. Automated detection aids radiologists in diagnosing intracranial aneurysms, thereby potentially expediting clinical workflows.
Binary Segmentation of Intracranial Aneurysms. We will provide the training set with 40 TOF-MRA volumes and corresponding binary annotations. Participants may use any other public datasets and private in-house data for training purpose. Test data includes 10 TOF-MRA volumes which is unavailable to the participants, and the participants will submit the models for performance evaluation.
The motivation of this Competition is to develop a system capable of autonomously segmenting intracranial aneurysms from TOF-MRA images. The incorporation of volumetric segmentation techniques enables comprehensive analysis of aneurysm size and shape, potentially yielding novel biomarkers crucial for refining rupture risk prediction models. Ultimately, the integration of such advancements into clinical practice has the potential to revolutionize the decision-making process surrounding the treatment of intracranial aneurysms, facilitating more informed and tailored therapeutic interventions. The overall aim is to develop an AI pipeline for Neuro-interventional Procedures.
Postgraduate Institute of Medical Education and Research is a public medical university in Chandigarh (PGIMER), INDIA.
A variety of Philips/Siemens MRI scanners (1.5 and 3T) were used to acquire the TOF-MRA Images. Due to the clinical nature of this data set and the fact that it was taken from multiple studies across many years, there is no set protocol or acquisition used across cases. This provides a diverse and realistic data set in which standard clinical protocols and routines were used.
Train Dataset: 40 Cases, Test Dataset: 10 Cases.
Training and test cases both represent a Time of Flight MRA of a human brain. Training and test cases will have corresponding annotations of the aneurysm based on the TOF-MRA to use for training or evaluation of the method. We are not providing a specific validation set and it is up to the participants to decide a validation/train set split based on the provided training data.
The IAs annotations are manually labelled voxel-wise by radiologist who have undergone comprehensive education of Cerebral anatomy, facilitated by clinical experts. Cases where annotators encounter uncertainty are flagged for additional verification and approval by the clinical expert team. This expert team comprises neuro-radiologists, neurologists and neurosurgeons ensuring a multidisciplinary approach to validation.
| May 1, 2024 | Website live and online |
| May 30, 2024 | Release of First set of training data |
| June 10, 2024 | Release of Second set of training data |
| June 27, 2024 | First deadline for submission of methods |
| July 10, 2024 | Final deadline for submission of methods |
The results will be announced publicly at the ICPR-2024 challenge session. After the session, the results will be published on the challenge website. A certificate will be provided to the top teams that have submitted before the deadline and are ranked in the top five for the task. The 1 st place team will be awarded $300, the 2 nd place team will receive $200, and the 3 rd place team will receive $100. A participation certificate will be provided to whosoever has submitted the methods before the deadline.
The training data will be available shortly.
The data distribution, registration and automatic evaluation will be handled by CIAS challenge team. The following link let you register into the challenge and await for the administrators confirmation in participating.
It is highly recommended to use your institutional email address for the registration. Each team cannot have more than five team members, and each participant can only join one team. Each team can only submit one algorithm for final ranking.
You will receive an email with an acceptance or decline in your team participating telling you the reasons. We cordially ask you for your patience while waiting for a response from CIAS team. Later on other mail, you will find an unique link to download the dataset. In case you didn't get to download the data, please send us an email to our email to generate a new link.
Click the following link to submit your method.
You will be able to upload your model contained in a zip file following the next steps:
In order to make a successful submission you can download and follow the comments instructions in the docker template zip which contains a Dockerfile and evaluate.py files with their respective instructions. The following are general rules to submit your solution in the challenges:
Your zip must contain the Dockerfile and evaluate.py which have defined elements that mustn't be modified in order to run.
You can attach inside the zip your model weights and model structure in any ".py", ".h5", ".pth", etc. There are no rules for the files contained in the zip file, except for Dockerfile and evaluate.py files.
In case of any error during the execution of the uploaded zip, the organizers will contact you via email.
Indication of how this metrics can be determined can be found here

Dr. Ashis Kumar Dhara
Main Organizer
Dept. of Electrical Engineering, NIT Durgapur INDIA.

Mr. Subhash Chandra Pal
Co-Organizer
PhD Student, Dept. of Electrical Engineering, NIT Durgapur INDIA.

Dr. Chirag Kamal Ahuja
Clinical Expert
Department of Radiodiagnosis and Imaging, PGIMER, Chandigarh, INDIA.

Prof. Johan Wikström
Clinical Expert
Department of Surgical Sciences, Neuroradiology, Uppsala University, SWEDEN.

Prof. Robin Strand
Technical Lead Advisor
Department of Information Technology, Vi3; Image Analysis, Uppsala University, SWEDEN.




Main Organiser:
Dr. Ashis Kumar Dhara,
Dept. of Electrical Engineering, NIT Durgapur INDIA.
E-Mail: akdhara.ee@nitdgp.ac.in
Co-Organiser:
Mr. Subhash Chandra Pal,
Dept. of Electrical Engineering, NIT Durgapur INDIA.
E-Mail: scp.21ee1109@phd.nitdgp.ac.in